In this section, experiments on real hyperspectral data are presented to evaluate the endmember extraction accuracy and parallel performance of the efficient implementation of ACOEE.
4.1. Computing Facilities and Dataset
The computer utilized in the experiments was equipped with two Intel Xeon E5-2620 CPUs, 128 GB RAM, and eight NVIDIA TITAN Xp GPUs.
Table 1 shows the features of the GPUs utilized in the experiments, which were connected to the computer using the PCI-Express 2.0 bus. The experiments were performed on the Ubuntu 16.04 operating system, in which the CUDA development environment 8.0 was installed. The proposed MG-ACOEE algorithm was carried out using CUDA C language, while O-ACOEE and G-ACOEE were carried out separately using MATLAB and CUDA C for comparison purposes.
Two popular hyperspectral datasets were adopted in the experiments for evaluating the performance of the proposed algorithm (see
Figure 4). The first one was the well-known Cuprite scene acquired by the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) [
40]. There were several minerals in this district, such as alunite, calcite, kaolinite, and muscovite. The scene utilized in this section comprised 400 × 350 pixels, and 50 bands ranging from 1.99–2.48 μm. The other one was the Urban data captured by Hyperspectral Digital Imagery Collection Experiment (HYDICE) in October 1995, which is one of the most widely-used hyperspectral datasets for hyperspectral unmixing research [
41,
42,
43]. The image is of size
, and there were 210 bands ranging from 400–2500 nm in each pixel. Due to dense water absorption and atmospheric effects, the bands 1–4, 76, 87, 101–111, 136–153, and 198–210 were removed, and 162 bands remained in these data. The main ground objects in this scene include asphalt road, grass, tree, and roof.
Aiming at reducing the size of feasible solution space, 80 pixels with the highest pixel purity index were extracted as candidate endmembers by the pixel purity index (PPI) algorithm [
6]. The number of endmembers in the Cuprite dataset was first estimated by HySime [
44], which provided an estimation of
endmembers. According to the literature [
41,
42], there are two versions of the ground truth of Urban data, which contain four and six endmembers, respectively. The number of endmembers in the Urban data was set to six in this paper. In order to reduce the computational complexity, 2000 pixels were uniformly sampled from the original hyperspectral datasets, instead of all pixels, for fully-constrained abundance inversions in ACOEE. The times in all the tables of this section are in seconds.
4.2. Endmember Extraction Accuracy and Parallel Computing Performance
The endmember extraction accuracy and parallel performance of O-ACOEE, G-ACOEE, and MG-ACOEE are compared in this section. The spectral angle distances and RMSE were adopted to evaluate endmember accuracy. Because ACOEE is a random search algorithm, the number of iterations varied with each run. Therefore, the computing performance was evaluated by the time per iteration (TPI). The mean and standard variances of the metrics over five runs for these algorithms were given in the experiments.
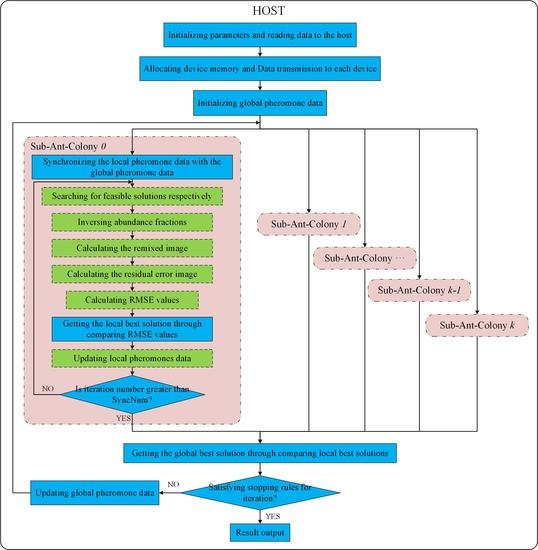
The ant number in these algorithms was set to 256. The number of sub-ant-colonies in MG-ACOEE, i.e., the number of GPUs, was eight. This means that the ant number in each sub-ant-colony of MG-ACOEE was 32. In MG-ACOEE, the number of iterations in a synchronous cycle, e.g., SyncNum, was set to four.
Four highly representative minerals (i.e., alunite, calcite, kaolinite, and muscovite) in the cuprite mining district were utilized for endmember accuracy comparison.
Table 2 and
Table 3 respectively report the spectral angle distances between USGS mineral spectra and their corresponding endmembers extracted by O-ACOEE, G-ACOEE, and MG-ACOEE with only ASC and full constraint. Asphalt road, grass, tree, and roof in the Urban data were considered for endmember extraction accuracy comparisons between the extracted endmembers and the ground truths, which are shown in
Table 4 and
Table 5. From the analyses of spectral angle distances and RMSE values in these tables, it could be found that the three algorithms all successfully extracted the considered endmembers in the experiments on Cuprite data and Urban data, and MG-ACOEE obtained comparable results, compared with O-ACOEE and G-ACOEE.
However, the computing performance of MG-ACOEE was superior to that of O-ACOEE and G-ACOEE in the experimental results. The total time, iteration numbers, and TPI of these three algorithms are reported in
Table 6,
Table 7,
Table 8 and
Table 9. It should be noted that the total time not only included the processing time on the host and the device, but also the time of data allocations and data transmissions. In these experiments, both the total time and TPI were greatly reduced in MG-ACOEE. Thanks to the multi-GPU features, TPIs of MG-ACOEE with only ASC or full constraint were respectively reduced 6.80- and 7.40-times for Cuprite data and 7.38- and 6.87-times for Urban data, when compared with G-ACOEE.
In summary, the parallel computing performance of MG-ACOEE was significantly improved under the premise of ensuring endmember accuracy.
4.3. Influence of Key Parameters
Key parameters in MG-ACOEE can affect the iteration number, TPI, and even the searching ability. In this subsection, more experiments of MG-ACOEE with only the ASC constraint were carried out in order to evaluate the influence of the number of GPUs (GPUsNum), the number of ants in a sub-ant-colony (AntsNum), and the number of iterations in a synchronous cycle (SyncNum).
The mean and standard variances of the RMSE, iteration number, total time, and TPI over five runs for these algorithms are given in the following experiments. For convenience, iteration number and total time were abbreviated to IN and TT.
1. Influence of GPUsNum:
In MG-ACOEE, the ant colony can be divided into more sub-ant-colonies as GPUsNum increases, if there is a certain number of ants in an ant colony. Four experiments were executed to evaluate the influence of the number of GPUs (GPUsNum), in which the number of ants in the ant colony was 256, and the GPUsNum was respectively set to 1, 2, 4, and 8. Obviously, it was G-ACOEE when the GPUsNum was equal to one. AntsNum was respectively 128, 64, and 32, when GPUsNum was 2, 4, and 8. SyncNum was set to four in these experiments.
Table 10 and
Table 11 show the RMSE, IN, TT, and TPI of these experiments separately on Cuprite and Urban data. From these tables, we can observe that the total time and times per iteration significantly decreased as
GPUsNum increased, while the RMSE was basically stable. This means that the computing performance of ACOEE can benefit from the proposed parallel strategy, in which multiple GPUs were utilized to accomplish the computing tasks of the subsidiary ant colonies in an ant colony.
Figure 5 reveals the speed ratios of MG-ACOEE with different
GPUsNum and G-ACOEE. When the
GPUsNum increased, the speed ratio correspondingly increased. However, the speed ratios could not come up to
GPUsNum, due to the time cost of data synchronization among different GPUs. It also can be found that the speed ratios in the experiments of Urban data were higher than the ones in the experiments of Cuprite data. That is because there were more bands in the Urban data, and more time was spent on searching for the optimal solution instead of data synchronization.
2. Influence of AntsNum:
If the the number of sub-ant-colonies (GPUsNum) was fixed, the number of ants per sub-ant-colony (AntsNum) not only determines the total number of the ant colony, but could also impact the parallel performance of MG-ACOEE. In this subsection, GPUsNum was set to eight, and five experiments were set up to evaluate the influence of AntsNum, in which the AntsNum was respectively set to 4, 12, 20, 28, and 32. The ant numbers in the colony correspondingly were 32, 96, 160, 224, and 256. SyncNum was set to four in these experiments.
Table 12 and
Table 13 report the average values and standard variances of RMSE, IN, TT, and TPI in these experiments. As
AntsNum increased from 4–32, the RSME value decreased from 3.052–3.018 in the experiments on Cuprite data, which meant that the searching ability of MG-ACOEE distinctly enhanced. Since there were more ants in the colony, the computing amount searching for optimal solution accordingly increased. If the TT and TPI of MG-ACOEE with
AntsNum = 4 was taken as a benchmark,
Figure 6 reveals the time ratios of MG-ACOEE with different
AntsNum and the benchmark. In this figure, it can be found that the TPI time ratios for Cuprite and Urban data, indicated by the
TPI(Cuprite) and
TPI(Urban) curves, were less than the
AntsNum ratios in the MG-ACOEE with different
AntsNum. For example, if
AntsNum = 32, the
AntsNum ratios was eight, while the TPI time ratios for Cuprite and Urban data were respectively 5.00 and 6.28. That was because the computing time searching for the optimal solution increased as
AntsNum increased, while the synchronizing time cost essentially was unchanged. This also means that the percentage of synchronizing time in TT decreased accordingly. For the same reason, the TPI time ratios on Cuprite data were less than the ones on Urban data. As a result of enhanced searching ability in an iteration, less iterations were needed in MG-ACOEE with more ants in the colony to obtain the optimal solution. Therefore, TT time ratios were even less than TPI time ratios for both Cuprite and Urban data, shown by the
TT(Cuprite) and
TT(Urban) curves.
3. Influence of SyncNum:
In MG-ACOEE, ants in a sub-ant-colony complete SyncNum iterations in a synchronous cycle, and then, the global best solution and global pheromone data are updated and the convergence conditions checked. SyncNum represents the frequency of synchronizing operations in MG-ACOEE, which are closely related to time per iteration and the convergence rate of the algorithm.
In this subsection, six experiments were carried out to evaluate the influence of
SyncNum.
GPUsNum and
AntsNum were separately set to eight and 32 in all the experiments, and
SyncNum was respectively set to 4, 8, 16, 32, 64, and 96. In the same way, the algorithms were run five times separately on Cuprite and Urban data, and then, the average values and standard variances of RMSE, IN, TT, and TPI are reported in the
Table 14 and
Table 15.
The variations of TPI and IN in Cuprite and Urban experimental results also can be found in
Figure 7. It was revealed that TPI slightly decreased, whereas IN considerably increased, when
SyncNum increased from 4–96. This was because the synchronizing number and correlative time cost was reduced accordingly. However, it should be particularly noted that there were fewer chances to check the convergence conditions of the algorithm as
SyncNum went up. Therefore, the convergence conditions were more difficult to satisfy, and IN notably increased by more than 80%; as a result, more time was spent on obtaining the optimal solutions. Thanks to the more full searching in the solution space, the RMSE values were slightly reduced in both the Cuprite and Urban experimental results.
4.4. Discussions
In the experiments on two real hyperspectral datasets, both endmember extraction accuracy and parallel performance were evaluated, and then, the influences of key parameters, i.e., GPUsNum, AntsNum, and SyncNum, were analyzed. For a colony with a fixed number of ants, the computing performance of MG-ACOEE was significantly improved owing to the multi-GPU parallel computing technology, when compared with O-ACOEE and G-ACOEE. Moreover, the advantage of MG-ACOEE on computing performance would be greater if more GPUs were utilized, while maintaining their endmember extraction accuracy. Therefore, it is proposed that GPUsNum, i.e., the number of sub-ant-colonies, should be set as big as possible in a multi-GPU computing system. If GPUsNum was fixed, total times and times per iteration did not increase linearly as AntsNum increased, because less time was utilized for data synchronization. It was a cost-efficient choice to set AntsNum = 32 in the MG-ACOEE for the reason that the RMSE of endmembers could be reduced owing to more ants used for searching for the optimal solution. In the experiments, SyncNum increasing could make the times per iteration slightly decrease and the iteration number considerably increase. Considering the lower RMSE values along with fuller searching in the solution space, SyncNum is recommended to be set to 32.
From the experimental analyses, it could be found that the RMSE values were closely related to the ant number in the colony and the iteration number. In other words, fuller searching in the solution space was conducive to obtaining higher precision endmember results. MG-ACOEE has shown high computational efficiency and has great potential to further improve the searching capability without too much time cost.