Abstract
When stress factors trigger transcriptional and metabolic changes, RNA interference (RNAi) is associated with gene expression regulation at the transcriptional and post-transcriptional levels. RDR, DCL and AGO proteins contribute to these gene silencing processes during stress reactions and plant development. An entire revision of the maize RDR, DCL and AGO genes was carried out prior to the experiments. In this study, the transcript changes of a total of 4 ZmRDR, 5 ZmDCL and 17 ZmAGO genes were analysed in maize during either drought stress or MDMV infection, with or without salicylic acid pre-treatment or siRNA pre-treatment, respectively. The gene expression profiles showed the early, middle and late activity of these genes. Drought stress caused major changes in the expression profiles, indicating that there were various steps in stress response regulation. Moreover, insights were gained into the fine-tuning mechanisms of SA regulation. In the case of MDMV infection less diverse trends were observed, which were mainly focused on antiviral defence. However, treatment with exogenous siRNA seems to be an appropriate tool for the targeted influencing of RNAi, especially of AGO genes. These results represent the first contribution to the relationship between RNAi and salicylate signalling and between viral infection and siRNA-triggered defence in maize.
Similar content being viewed by others
Introduction
Maize is one of the most widely grown cereals worldwide. The world's total maize production was estimated at 1.05 million thousand tonnes in 2019 (Knoema 2019). It has very diverse uses: in addition to animal feed, its industrial processing is also important for the production of starch, invert sugar and alcohol. Furthermore, maize is a staple food in the most populous, poorer regions of the world. This important crop plant is exposed to many abiotic and biotic stress factors in the fields. Not only can the highly variable and often unpredictable weather test plant defence systems, but severe abiotic stress may further increase the susceptibility of plants to certain biotic stressors (Trębicki and Finlay 2019).
Drought, extreme temperature changes, heavy metals, increased soil salt content and certain pathogens (fungi, bacteria, viruses, insects) are able to launch a finely tuned signalling (e.g. MAPK—mitogen activated protein kinase, transcription factors) and gene expression system that elicits stressor-specific metabolic responses, enabling plants to survive (Atkinson and Urwin 2012). Another important regulatory mechanism is also involved in the transcriptional and post-transcriptional regulation of these gene networks, namely RNA interference (RNAi). RNAi is an evolutionarily conserved defence mechanism that allows sequence-specific cleavage following the recognition/formation of endogenous or exogenous double-stranded RNA molecules. The key proteins involved in the maturation and formation of these molecules are RNA-dependent RNA polymerases (RDRs), Dicer-like nucleases (DCL) and Argonaute RNA-binding proteins (AGO) (Shabalina and Koonin 2008).
The 20–30 nt long microRNA (miRNA) molecules endogenously generated in the process of RNAi are principally responsible for plant development, the regulation of metabolism, the maintenance of genome integrity, and even the development of stress responses. These miRNA molecules specifically bind to the AGO-RISC (Argonaute—RNA-induced silencing complex) complexes containing the corresponding AGO proteins, thereby contributing to the sequence-specific binding and cleavage of transcripts of the genes to be silenced (Iwakawa and Tomari 2015; Gan et al. 2017). In addition to post-transcriptional gene silencing, some miRNAs induce chromatin rearrangement by maintaining DNA methylation, thereby inhibiting transcription from their target gene (Axtell 2013; Achkar et al. 2016). Accordingly, DCL, RDR and AGO isoenzymes are involved in numerous functions and affecting, among other things, the type and intensity of stress response and plant development (Fang and Qi 2016; Liu et al. 2018).
Salicylic acid (SA) is a key plant growth regulator influencing developmental processes and plant immunity. It has an important regulatory role in stress responses to both biotic and abiotic challenges. During the development of a given stress response, RNAi participates in the fine-tuning of the classical sense signalling pathway, with the involvement of small RNA (sRNA) molecules (Samad et al. 2017). However, the role of RNAi genes has not yet been clarified in the case of drought stress and SA signalling.
Among the biotic stressors that threaten sweet corn, plant pathogens, including viruses, play the main role. Maize dwarf mosaic virus (MDMV) pathogen uses plant resources to ensure its own intracellular growth, thereby contributing significantly to slowing down the development and growth processes of the plants (Kannan et al. 2018). In the case of MDMV infection, the single stranded RNA genome released from the coat protein units in the cytoplasm serves as the signal to the plant and the RNAi process is activated almost immediately. Small interfering RNA (siRNA) copies of the viral genome are rapidly formed due to the activity of the RDR and DCL enzymes, and then selectively incorporated into the AGO-containing RISC complexes. The AGO protein then mediates the sequence-specific binding and catalytic cleavage of the viral RNA designated by the sRNA molecule (Llave 2010; Khraiwesh et al. 2012). Utilizing the working principle of RNAi, plant defence can be activated by treatment with longer double-stranded RNA molecules (dsRNA) or shorter small interfering RNA duplexes, mimicking the infection, but without real danger. Therefore, it may ultimately be possible to prepare the plants for infection, thus contributing to slower viral proliferation or even complete inhibition (Konakalla et al. 2016; Kaldis et al. 2018).
The strength and effectiveness of the plant stress response significantly depend on the regulatory steps. Obtaining a better understanding of the gene expression patterns of sRNA molecules and RNA-interfering enzymes and of their relationships would offer numerous hitherto untapped opportunities to enhance the stress response and to alleviate the damage caused by stress factors. Thus, exploring the regulation of plant stress responses could provide information that would significantly improve the stress capacity of crops (Kamthan et al 2015).
The aim of the present work was to identify RNAi-associated genes in maize plants, to clarify their function and to characterise the expressional changes and dynamics of the system under (i) drought stress with or without salicylic acid pre-treatment and (ii) MDMV infection with or without siRNA pre-treatment.
Materials and Methods
Identification of DCL, RDR and AGO Genes
Maize genome and transcript sequences were downloaded from MaizeGDB (https://www.maizegdb.org/) and Gramene (https://www.gramene.org/). The downloaded sequences can be found in the supplementary files AGOcDNA.txt, DCLcDNA.txt and RDRcDNA.txt. Hidden Markov Model analysis and BLAST search alignment tools (Cannon et al. 2011) were used to find the DCL, RDR and AGO genes of the B73 maize genome reference version 5.0 of MaizeGDB. The blastn algorithm of the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/) was used to find the homologous and similar sequences encoded by the Arabidopsis thaliana and Oryza sativa genomes, aiming to identify the corresponding genes in the B73 maize genome. The Clustal Omega multiple sequence alignment program (https://www.ebi.ac.uk/Tools/msa/clustalo/) was used for the alignment analyses of similar sequences in a given gene family or subfamily. Phylogenetic analysis was carried out with the MEGA X program (Kumar et al. 2018) using the neighbor-joining method with 1,000 bootstrap replicates. Genome Browser tool of MaizeGDB, optimised for reference version 5.0 (https://jbrowse.maizegdb.org/), was used for mapping the chromosomal localisation of the DCL, RDR and AGO genes of maize.
Plant Material, Growth Conditions and Treatments
For the drought experiment, maize plants (Zea mays L. cv. MV 350) were grown in soil in a greenhouse with controlled climate and light conditions (14/10-h light/dark period, photosynthetic photon flux density (PPFD) of 200 μmol m−2 s−1, 25 °C and ~ 40% relative humidity). Three-day-old germinated kernels were transferred to a 70% water capacity soil:sand (2:1) mixture, which was checked and moistened every day with 1⁄4 strength Hoagland solution (containing 80 μM Fe(III)-EDTA as iron form). Salicylic acid (SA) treatment was carried out by spraying 3 ml of 0.1 mM SA solution and 0.00025% Nonit mixture on each 11-day-old plant. Non-stressed and drought-stressed plants were sprayed with 3 ml 0.00025% Nonit solution. The plants were exposed to 9-day drought stress from the age of 12 days. Samples were taken 3, 6 and 9 days past the beginning of drought (dpd). Drought conditions were controlled through the gradual reduction of soil water capacity from 70 to 50% (Fig S1). The sample plants were separated into 4 groups: DW—non-stressed control plants; SA—SA-pretreated plants; DW-D—plants exposed to drought stress; SA-D—SA-pretreated plants exposed to drought stress (Fig. S2). The appropriate treatment dose, drought conditions and sampling dates were determined during preliminary experiments based on physiological measurements.
Sweet corn (Zea mays cv. Saccharata var. Honey Koern.) was used to study the mechanism of RNA interference under biotic stress. Pre-germinated corn grains were grown hydroponically, on ¼ strength Hoagland solution (containing 80 μM Fe(III)-EDTA as iron form). The plants were grown at 250 μmol m−2 s−1 PPFD, 23/25 °C temperature and 50% relative humidity, in a SANYO MLR-350 HT (SANYO Electric Co., Ltd., Japan) plant growth chamber with a 14/10-h light/dark period. Plants without subsequent treatments were indicated as control (CO) plants (Fig S3). To investigate the effect of sRNA treatment on RNA interference, 10-day-old plants were treated with siRNA molecules (treatment group siRNA). This siRNA sequence (Sense sequence: 5-P.G.A.A.G.C.A.C.A.G.A.A.G.G.A.G.G.C.A.G.A.G; Antisense sequence: 5-P.C.U.C.U.G.C.C.u.c.c.U.U.C.U.G.U.G.C.U.U.C) is complementary to the 5' portion of the coat protein subunit coding region of the MDMV genome and was determined in a previous sRNA sequencing project. 10 μl of 30 ng/μl siRNA solution was injected into the open leaf sheaths of the maize plants. 11 and 13 days after the germination the plants in the MDMV group were infected mechanically with the Dallas A strain of MDMV. 1 g leaf tissue from infected plants developing macroscopic symptoms were homogenized in 10 ml Sörensen phosphate buffer (pH 7.2, 0.06 M) and were used for inoculating the first and second leaves of MDMV group plants. Carborundum was added as an abrasive. To investigate the effects of siRNA pretreatment in infected plants, siRNA-pre-treated plants were infected with the MDMV Dallas A strain (henceforth referred to as the siRNA-MDMV group). Sampling was performed one, two, and three weeks past the first MDMV infection (wpi).
Analysis of Gene Expression
Total RNA samples were isolated from the second leaves of maize plants using the Direct-zol RNA Miniprep Kit (Thermo Fisher Scientific, Rockford, IL, USA), including the DNA digestion step. cDNA was synthesised from 500 ng RNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). qRT-PCR reactions were run on an ABI StepOnePlus Real-Time PCR instrument (Thermo Fisher Scientific), using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific). Folylpolyglutamate synthase (FPGS), leunig (LUG) and membrane protein PB1A10.07c gene (MEP) were used as internal control genes to normalise the Cq values of the genes of interest (Manoli et al. 2012). The geometric mean of the internal control data across biological and technical replicates were applied for normalisation. The relative changes in gene expression were compared to the untreated control group and quantified according to the Pfaffl method (Pfaffl 2004). Primers were designed by Primer3 online software (Koressaar and Remm, 2007) and fine-tuned manually when necessary. Reaction efficiencies were calculated via the LinRegPCR software (Ramakers et al. 2003). The expression of the following genes was analysed by qRT-PCR: Dicer-like 1, 2, 3a, 3b, 4 (DCL1, DCL2, DCL3a, DCL3b, DCL4); Mediator of paramutation 1 (MOP1); RNA-dependent RNA polymerase 1, 4, 6 (RDR1, RDR4, RDR6); Argonaute 1a, 1b, 1c, 1e, 2b, 4a, 4b, 4c, 5a, 5b, 6, 7, 10a, 10b, 18a, 18b (AGO1a, AGO1b, AGO1c, AGO1e, AGO2b, AGO4a, AGO4b, AGO4c, AGO5a, AGO5b, AGO6, AGO7, AGO10a, AGO10b, AGO18a, AGO18b) (Table S1).
Data Evaluation and Statistical Analysis
Three technical repeats and three biological repeats were used for the qRT-PCR experiments. After checking the normality of the data the results were statistically evaluated with ANOVA and Tukey’s honest significant difference (TukeyHSD) post-hoc test at the 5% significance level (p ≤ 0.05) using the RStudio program package (Racine 2012). The heatmaps of the centred and scaled log2 relative expression values were generated by the pheatmap package in RStudio (https://CRAN.R-project.org/package=pheatmap). The gene clusters were classified by Z-score determination of each row. The data used for the analyses can be found in the supplementary table (Table S2).
Results
DCL, RDR and AGO Genes in Maize
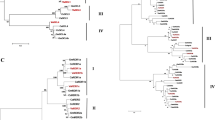
Qian et al. (2011) identified 5 DCL, 5 RDR and 18 AGO genes in the maize genome. Zhai et al. (2014) described 17 AGO sequences, 12 of which were among the 18 AGO gene predictions of Qian et al. (2011) and five more. The present gene hunting project approved the DCL genes (Table 1) identified by Qian et al. (2011), but it was proposed that the earlier data should be reconsidered for the RDR and AGO genes. Seven RDR genes (Table 1) and 20 AGO genes (Table 2) were distinguished in maize in the present work, some of them not found earlier and some described under another name. All the gene names given by Qian et al. (2011) were affirmed, except those for which revision was indispensable. For instance, ZmRDR4 has been reconsidered as the former description was identical to the recently identified ZmRDR6, while a new gene with the name ZmRDR4 has been annotated here as an AtRDR3/4/5-OsRDR3/4 type gene and named after its similarity to OsRDR4. The ZmRDR5 gene described by Qian et al. (2011) has been renamed ZmRDR5a, as two closely related genes were found in the maize genome (Fig. 1). The AGO genes were revised, first according to their similarity to the corresponding rice genes and secondly to well-known genes in Arabidopsis (Fig. 2).
Phylogenetic relationships between RDR transcripts of maize, rice and Arabidopsis. Bootstrap values larger than 50 out of 1000 bootstrap replications are shown for each clade on the unrooted neighbour-joining (NJ) tree. Accession numbers and abbreviations: OsRDR1 (Os02g50330), OsRDR2 (Os04g39160), OsRDR3 (Os01g 10130), OsRDR4 (Os01g10140) and OsSHL2 (Os01g34350); AtRDR1 (At1g14790), AtRDR2 (At4g11 130), AtRDR3 (At2g19910), AtRDR4 (At2g19920), AtRDR5 (At2g19930) and AtRDR6 (At3g49500)
Phylogenetic relationships between AGO transcripts of maize, rice and Arabidopsis. Bootstrap values larger than 50 out of 1000 bootstrap replications are shown for each clade on the unrooted neighbour-joining (NJ) tree. Accession numbers and abbreviations: OsAGO1a (Os02g45070), OsAGO1b (Os04g47870), OsAGO1c (Os02g58490), OsAGO1d (Os06g51310), OsAGO2 (Os04g52540), OsAGO3 (Os04g52550), OsAGO4a (Os01g16870), OsAGO4b (Os04g06770), OsAGO14 (Os07g09020), OsMEL1 (Os03g58600), OsAGO17 (Os02g07310), OsAGO12 (Os03g47820), OsAGO18 (Os07g28850); AtAGO1 (At1g48410), AtAGO2 (At1g31280), AtAGO3 (At1g31290), AtAGO4 (At2g27040), AtAGO5 (At2g27880), AtAGO6 (At2g32940), AtAGO7 (At1g69440), AtAGO8 (At5g21030), AtAGO9 (At5g21150) and AtAGO10 (At5g43810)
Expression Patterns of RNAi Genes During SA-Treatment and Drought Stress
Analysis of the gene expression profiles revealed early, middle and late activated RNAi genes. The gene activities and activity changes corresponded to mild, moderate and strong stress conditions, related to the decrease in field water capacity resulting in drought stress. According to the expression data, various trends could be observed in the drought-stressed groups. The expression of DCL1, DCL2, AGO2b, AGO7 and AGO18b showed a gradual increase throughout the experiment from inhibited to enhanced gene activity. DCL3b, RDR1, AGO1c, AGO4b, AGO10a and AGO10b showed a gradual decrease from enhanced to inhibited activity in both drought-stressed groups. Drought stress caused an “A”-shaped trend in the case of DCL3a, DCL4, MOP1, RDR6, AGO1a, AGO2a, AGO4a and AGO5b and a”V”-shaped pattern for AGO1b, AGO5a and AGO18a. The gradually increasing and decreasing expression patterns may indicate the early and late importance of these genes, respectively, the V-shaped pattern showing a dual early-late role and the A-shaped trend corresponding to middle-activated genes (Fig. 3).
The gene expression heatmap visualizes the difference between expression data over the time course of each treatment. Genes with similar expression patterns form a cluster, so basically three groups that may play a major role in drought stress responses can be distinguished (Fig. 4). DCL3b, RDR1, AGO4b and AGO5a were early activated genes, whose activity decreased over time to the level of the DW group or became completely suppressed. The expression of these genes was also affected by SA pretreatment. MOP1 had constant high activity in all the treatments, with an expression peak at 6 dpd for DW-D and SA-D. The activity of this gene remained high in the case of DW-D, while in SA-D there was a slight decrease. DCL4 and AGO18b were activated later, but their expression remained high till the end of the treatment in DW-D and SA-D. DCL1, DCL2, AGO2b and AGO7 were late activated genes. Apart from AGO18b, AGO7 had the largest change in expression, indicating its importance in the late phase drought stress response. SA pre-treatment caused mostly minor changes, though RDR1, AGO4a and AGO5a showed early, DCL4 and AGO5b middle and AGO7 late responses, while MOP1 had constant high activity.
Cluster analysis of gene expression based on the log2 relative expression data during SA treatment and drought stress. The columns show the gene clusters and the rows the experimental conditions at a given time. The gene clusters were classified by the Z-score determination of each row. Red indicates up-regulated and blue down-regulated genes
Expression Pattern of RNAi Genes During MDMV Infection
The expression activity of RNA interference genes examined during the first three weeks of an MDMV infection showed several changes. As a result of the infection, increasing expression activity was observed in the RDR and DCL gene clusters during the first week, which mainly affected DCL3a and RDR1 (Fig. 5). However, gene activation was the most expressed in the AGO cluster, affecting a total of 6 genes (AGO1a, AGO1e, AGO10a, AGO10b and AGO18a). Thus, together with other stress-responsive genes, these RNAi genes may play an important role in the development of the early stress response during MDMV infection. In most cases expression decreased during the second and third week of infection, resulting in a gradually decreasing or V-shaped trend (Fig. 5).
Genes showing different expression activity trends can be clearly distinguished on the heatmap (Fig. 6). The gene expression trends of AGO1a, AGO4b, AGO10a, AGO10b, RDR1 and DCL3a were similar and appeared to be activated early in each treatment. In the case of AGO1a, AGO4b, and RDR1 this activation extended into the second week as well. The following group, consisting of AGO1e, AGO2a and AGO2b, exhibited increased activity to a greater or lesser extent in all three weeks during the course of infection. The only late activated genes were AGO7 and AGO18b. According to the heatmap, the activity of AGO18a was clearly the most prominent, showing its key role in the defence against MDMV in this maize variety (Fig. 6).
Cluster analysis of gene expression based on the log2 relative expression data during siRNA treatment and MDMV infection. The columns show the gene clusters and the rows the experimental conditions at a given time. The gene clusters were classified by the Z-score determination of each row. Red indicates up-regulated blue down-regulated genes
Discussion
The function of RNAi genes have been examined in a number of studies. Based on the available literature, in the present discussion we highlight the processes, regulatory steps and physiological changes in which genes that are activated in different periods, may be involved.
RNAi Expression Profiles can be Well Characterised During Drought Stress
RDR1, DCL3b, AGO4b and AGO5a proved to be early activated genes. As they regulate miRNA genesis and RNA-dependent DNA methylation (RdDM), the early stress responses at the beginning of drought stress involve both transcriptional and post-transcriptional gene regulation (Axtell 2013). MOP1, RDR6, DCL3a, DCL4, AGO1a, AGO2a, AGO4a and AGO5b reached their expression peak in the middle of the 9-day stress experiment. In maize, the RDR2 homologue MOP1 gene is required for the establishment and maintenance of paramutations and transcriptional silencing. Moreover, the MOP1/DCL3 and RDR6/DCL4 siRNA pathways were reported to maintain the production of 22 nt siRNAs that take part in RdDM (Alleman et al. 2006; Nobuta et al. 2008). Jiang et al. (2020) found synergy between the RDR6/DCL4 siRNA pathway and the regulation of the carbon metabolism and anthocyanin biosynthesis in Arabidopsis. The present results suggest that SA pre-treatment enhanced the expression of RDR6 3 and 6 days after the beginning of drought. While in DW-D the initial activity constantly decreased, there was an expression peak at 6 days in the SA and SA-D groups. SA is known to enhance anthocyanin levels through its regulatory and direct metabolic effects (Khan et al. 2015), so RDR6 may be involved in this regulation. Two of the most important late activated genes were AGO7 and AGO18b. The product of AGO7 colocalizes and interacts with RDR6 and DCL4 enzymes and accumulates in siRNA bodies during stress (Jouannet et al. 2012). AGO7 is loaded with ta-siRNAs targeting auxin response factors (ARF3 and ARF4), thus taking part in organ development (Fahlgren et al. 2006; Jouannet et al. 2012). AGO18b is a negative regulator of the inflorescence meristem and shoot apical meristem in either the vegetative or the reproductive phase or during the presence of a stress factor (Wu et al. 2017; Sun et al. 2019). According to these data, AGO7 and AGO18b seem to be involved in regulating the balance between stress response and development. In this case the stronger the drought stress, the stronger the gene activity, which induces a gradual increase in the inhibition of SAM development, causing the growth of the seedlings to come to a halt during the presence of the stress factor.
RNAi is Involved in the Abiotic Stress Regulation of SA
From the abiotic stress point of view SA is a plant hormone inducing systemic changes while cooperating with other plant hormones (hormone crosstalk), so it is difficult to distinguish specific SA effects (Peleg and Blumwald 2011). The SA effect is well described according to biotic stress responses. However, numerous physiological, metabolomical and gene expression changes can be observed during abiotic stresses, as well. For example, the enhancement of photosynthetic efficiency and antioxidative enzyme activity or an increased level of polyamines are indirect effects resulting from regulatory changes caused by SA (Hayat et al. 2010). Moreover, stress protective SA treatment is concentration-dependent, suggesting the existence of fine-tuning mechanisms in the SA regulatory pathway, which has not been fully explored in terms of abiotic stress responses (Curaba et al. 2014; Hernández et al. 2017). Gene expression results show that, after perceiving the presence of the stress factor, plants respond with chromatin remodelling in order to induce stress response mechanisms, in which RNAi and RdDM have a crucial role (Zhang et al. 2018). Exogenous SA treatment causes an expression pattern similar to that of drought stress in several RNAi genes. Thus, according to the previously described role of RNAi proteins SA seems to induce a priming mechanism through RdDM and chromatin remodelling, which may result in the faster activation of stress-responsive metabolic routes, such as anthocyanin biosynthesis. This raises the question of whether the concentration-dependent effect of SA depends on the fine-tuning effect of siRNA pathways during abiotic stress responses.
Effects of MDMV Infection and siRNA-(pre)Treatments on RNAi Expression Profiles
Previously we examined this cultivar in detail in case of MDMV infection and various treatments. This cultivar shows great susceptibility for this type of virus and develops specific, well characterised symptoms. The MDMV can cause major changes in physiological, metabolomical and gene expression levels (Ludmerszki et al. 2017). However, RNAi expression patterns have not been studied yet in details.
As in the drought stress response, genes activated early or later in the process of RNA interference can be distinguished in the case of viral infection. Plant viruses, including MDMV use the plants resources to replicate their own genetic material and proteins, so that the plants lack the necessary resources for development and growth (Revers and Gracía 2015). Among other things, this results in the activation of gene silencing mechanisms at the post-transcriptional level and in chromatin modification, besides further antiviral defence mechanisms. This is also evidenced by a slight increase in the activity of genes that also showed a significant change in the case of drought. RNAi has an essential role in antiviral defence (Li et al. 2016). Besides its direct antiviral protection, RNAi also plays a role in metabolic and developmental changes during biotic stress. During viral infections, the appearance of exogenous viral RNA in the cytoplasm of the plant cell serves as a signal to the plant that activates the mechanism of RNA interference. Proteins encoded by the RDR1 and RDR6 genes are thought to be responsible for the formation of double-stranded RNA, which can then be diced to secondary siRNA molecules during geminiviral infection (Guo et al. 2018). In a further step in the process, 21–24 nt RNA molecules cleaved by DCL4 endonuclease proteins may be incorporated into AGO-RISC complexes. In the case of an RNA virus infection, AGO1 and AGO7 proteins are primarily responsible for the binding of these siRNA molecules (Szittya and Burgyán 2013). The present results suggest that RNA-dependent RNA polymerase encoded by RDR1 may have been responsible for the formation of viral siRNAs during MDMV infection. While there was no significant increase in the activity of the DCL2 and DCL4 genes, the expression of DCL3a showed a slight increase in the first week of the treatments. Regarding the activation of AGO genes, it can be stated that in this experimental system the increase in AGO18a activity was the most dominant, while only minor changes could be observed for the AGO1 and AGO7 genes. AGO18a is a monocot-specific gene, whose expression responds to viral infections and plays a major role in the antiviral defence of infected tissues (Wu et al. 2015). The results presented here show that the treatment with viral-origin siRNA also enhanced AGO18a expression, leading to high activity similar to that in virus-infected plants. For the other genes, the main activity changes were detected in response to MDMV infection. However, DCL3a, MOP1 and AGO4b had a higher expression rate at the first sampling date in the siRNA-MDMV group, indicating a stronger initial stress response against infection. These genes also exhibited enhanced activity in the siRNA group, suggesting that siRNA treatment activated a priming mechanism in the antiviral defence system. The late activation of AGO7 and AGO18b suggested the downregulation of growth, reflecting the trend seen in the abiotic stress response. The results thus demonstrate that exogenous siRNA application influences the basic expression pattern of RNAi genes, so targeted treatment could induce a priming mechanism in plants, enhancing the antiviral RNAi defence system before the appearance of a pathogen.
Conclusions
Stress factors trigger transcriptional and metabolic changes, in which RNAi is associated with gene expression regulation at the post-transcriptional level and with chromatin modification in transcriptional silencing. Moreover, it plays a major role in antiviral defence (Wendte and Pikaard 2016; Zhang et al. 2018). RDRs DCLs and AGOs contribute to these gene regulation processes during plant development as well (Finnegan and Matzke 2003). In this study, the activity of a total of 4 ZmRDR, 5 ZmDCL, and 17 ZmAGO genes were analysed during drought stress and MDMV infection in maize. Similar priming treatments were applied to reveal regulatory changes in maize during abiotic and biotic stress. The results revealed the differential expression patterns exhibited by these genes under abiotic and biotic stress conditions. The gene expression profiles showed the early, middle and late activation of these genes. Drought stress caused major changes in the expression profiles, indicating the stress response regulation takes place in several steps. Moreover, insights were gained into the fine-tuning mechanisms of SA regulation. We observed that salicylate signalling might be in relationship with RNAi. In the case of MDMV infection less diverse trends were observed, which were mainly focused on antiviral defence. However, treatment with exogenous siRNA seems to be an appropriate tool for the targeted influencing of RNAi, especially of AGO genes.
Availability of data and material
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Code availability
All software applied in this study is cited in this published article.
References
Achkar PN, Cambiagno AD, Manavella AP (2016) miRNA biogenesis: a dynamic pathway. Trends Plant Sci 12:1030–1044. https://doi.org/10.1016/j.tplants.2016.09.003
Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler EJ, White J, Sikkink K, Chandler V (2006) An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442:295–298. https://doi.org/10.1038/nature04884
Atkinson JN, Urwin EP (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63(10):3523–3544. https://doi.org/10.1093/jxb/ers100
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159. https://doi.org/10.1146/annurev-arplant-050312-120043
Cannon EKS, Birkett SM, Braun BL, Kodavali S et al (2011) POPcorn: an online resource providing access to distributed and diverse maize project data. Int J Plant Genom. Article ID 923035. https://doi.org/10.1155/2011/923035
Curaba J, Singh BM, Bhalla LP (2014) MiRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 65(6):1425–1428. https://doi.org/10.1093/jxb/eru002
Fahlgren N, Montgomery AT, Howell GM, Allen E, Dvorak KS, Alexander LA, Carrington CJ (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944. https://doi.org/10.1016/j.cub.2006.03.065
Fang X, Qi Y (2016) RNAi in plants: an argonaute-centered view. Plant Cell 25(2):272–285. https://doi.org/10.1105/tpc.15.00920
Finnegan EJ, Matzke MA (2003) The small RNA world. J Cell Sci 116:4689–4693. https://doi.org/10.1242/jcs.00838
Gan D, Zhan M, Yang F, Zhang Q, Hu K, Xu W, Lu Q, Zhang L, Lang D (2017) Expression analysis of argonaute, Dicer-like, and RNA-dependent RNA polymerase genes in cucumber (Cucumis sativus L.) in response to abiotic stress. J Genet 96:235–249. https://doi.org/10.1007/s12041-017-0758-y
Guo Z, Li Y, Ding SW (2018) Small RNA-based antimicrobial immunity. Nat Rev Immunol 19:31–44. https://doi.org/10.1038/s41577-018-0071-x
Hayat Q, Hayat S, Irfan M, Ahmad A (2010) Effect of exogenous salicylic acid under changing environment: a review. Environ Exp Bot 68:14–25. https://doi.org/10.1016/j.envexpbot.2009.08.005
Hernández AJ, Vivancos-Diaz P, Barba-Espín G, Clemente-Moreno JM (2017) On the role of salicylic acid in plant responses to environmental stresses. In: Salicylic acid: a multifaceted hormone. Springer, Berlin, pp 17–34. https://doi.org/10.1007/978-981-10-6068-7_2
Iwakawa H, Tomari Y (2015) The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol 25(11):651–665. https://doi.org/10.1016/j.tcb.2015.07.011
Jiang N, Gutierrez-Diaz A, Mukundi E, Lee SY, Meyers CB, Otegui SM, Grotewold E (2020) Synergy between the anthocyanin and RDR6/SGS3/DCL4 siRNA pathways expose hidden features of Arabidopsis carbon metabolism. Nat Commun 11(1):1–13. https://doi.org/10.1038/s41467-020-16289-3
Jouannet V, Moreno BA, Elmayan T, Vaucheret H, Crespi DM, Maizel A (2012) Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J 31:1704–1713. https://doi.org/10.1038/emboj.2012.20
Kaldis A, Berbati M, Melita O, Reppa C, Holeva M, Otten P, Voloudakis A (2018) Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol Plant Pathol 19(4):883–895. https://doi.org/10.1111/mpp.12572
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci 6:1–17. https://doi.org/10.3389/fpls.2015.00208
Kannan M, Ismail I, Bunawan H (2018) Maize dwarf mosaic virus: from genome to disease management. Viruses 10(9):492. https://doi.org/10.3390/v10090492
Khan RIM, Fatma M, Per ST, Anjum AN, Khan AN (2015) Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front Plant Sci 6:462. https://doi.org/10.3389/fpls.2015.00462
Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimi Biophys Acta Gene Regul Mech 1819 2:137–148. https://doi.org/10.1016/j.bbagrm.2011.05.001
Knoema (2019) Maize production quantity. https://knoema.com/atlas/topics/Agriculture/Crops-Production-Quantity-tonnes/Maize-production
Konakalla NC, Kaldis A, Berbati M, Masarapu H, Voloudakis AE (2016) Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 244(4):961–969. https://doi.org/10.1007/s00425-016-2567-6
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. https://doi.org/10.1093/bioinformatics/btm091
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Li M, Weng K, Shih S, Brewer G (2016) The evolving world of small RNAs from RNA viruses. Wiley Interdiscip Rev WIREs: RNA 7(5):575–588. https://doi.org/10.1002/wrna.1351
Liu H, Yu H, Tang G, Huang T (2018) Small but powerful: function of microRNAs in plant development. Plant Cell Rep 37(3):515–528. https://doi.org/10.1007/s00299-017-2246-5
Llave CÃ (2010) Virus-derived small interfering RNAs at the core of plant—virus interactions. Trends Plant Sci 15(12):701–707. https://doi.org/10.1016/j.tplants.2010.09.001
Ludmerszki E, Chounramany S, Oláh C, Kátay G, Rácz I, Almási A, Solti Á, Bélai I, Rudnóy S (2017) Protective role of S-methylmethionine-salicylate in maize plants infected with Maize dwarf mosaic virus. Eur J Plant Pathol. https://doi.org/10.1007/s10658-017-1174-0
Manoli A, Sturaro A, Trevisian S, Quaggiotti A, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol. https://doi.org/10.1016/j.jplph.2012.01.019
Nobuta K, Lu C, Shrisvastava R, Pillay M, Paoli DE, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong H-D, Yen Y, Green JP, Chandler LV, Meyers CB (2008) Distinct size distribution of endogenous siRNAs in maize: evidence from deep sequencing in the mop1-1 mutant. PNAS 105(39):14958–14963. https://doi.org/10.1073/pnas.0808066105
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295. https://doi.org/10.1016/j.pbi.2011.02.001
Pfaffl MW (2004) Quantification strategies in real time PCR. IUL biotechnology series; 5. In: Bustin SA (ed) A-Z of quantitative PCR. International University Line, La Jolla, pp 87–112
Qian Y, Cheng Y, Cheng X, Jiang H, Zhu S, Cheng B (2011) Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep 30:1347–1363. https://doi.org/10.1007/s00299-011-1046-6
Racine JS (2012) RStudio: a platform-independent IDE for R and Sweave. J Appl Economet 27:167–172. https://doi.org/10.1002/jae.1278
Ramakers C, Ruijter JM, Lekanne RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339(1):62–66. https://doi.org/10.1016/S0304-3940(02)01423-4
Revers F, García JA (2015) Molecular biology of potyviruses. Adv Virus Res 92:101–199. https://doi.org/10.1016/bs.aivir.2014.11.006
Samad FA, Sajad M, Nazaruddin N, Fauzi AI, Murad AMA, Zainal Z, Ismail I (2017) MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci 8:1–18. https://doi.org/10.3389/fpls.2017.00565
Shabalina AS, Koonin VE (2008) Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 21(10):578–587. https://doi.org/10.1016/j.tree.2008.06.005
Sun W, Chen D, Xue Y, Zhai L, Zhang D, Cao Z, Liu L, Cheng C, Zhang Y, Zhang Z (2019) Genome-wide identification of AGO18b-bound miRNAs and phasiRNAs in maize by cRIP-seq. BMC Genom 20:656. https://doi.org/10.1186/s12864-019-6028-z
Szittya G, Burgyán J (2013) RNA interference-mediated intrinsic antiviral immunity in plants. In: Cullen B (ed) Intrinsic immunity current topics in microbiology and immunology, vol 371. Springer, Berlin. https://doi.org/10.1007/978-3-642-37765-5_6
Trębicki P, Finlay K (2019) Pests and diseases under climate change; its threat to food security. In: Food security and climate change. ISBN: 978-1-119-18064-7, pp 229–241
Wendte MJ, Pikaard SC (2016) The RNAs of RNA-directed DNA methylation. BBA Gene Regul Mech 1:140–148. https://doi.org/10.1016/j.bbagrm.2016.08.00
Wu J, Yang Z, Wang Y, Zeng L, Ye R, Ji Y, Zhao S, Ji S, Liu R, Xu L, Zheng H, Zhou Y, Zhang X, Cao X, Xie L, Wu Z (2015) Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. Elife 4:1–19. https://doi.org/10.7554/eLife.05733
Wu J, Yang R, Yang Z, Yao S, Zhao S, Wang Y, Li P, Song X, Zhou T, Lan Y, Xie L, Zhou X, Chu C, Qi Y, Cao X (2017) ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants 3:16203. https://doi.org/10.1038/nplants.2016.203
Zhai L, Sun W, Zhang K, Jia H, Liu L, Liu Z, Teng F, Zhang Z (2014) Identification and characterization of Argonaute gene family and meiosis- enriched Argonaute during sporogenesis in maize. J Integr Plant Biol 56:1042–1052. https://doi.org/10.1111/jipb.12205
Zhang H, Lang Z, Zhu J-K (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19(8):489–506. https://doi.org/10.1038/s41580-018-0016-z
Acknowledgements
The authors wish to thank Eötvös Loránd University and the National Research, Development and Innovation Office (ÚNKP-19-3; GINOP-2.3.2) for funding the collection of materials, data analysis, and experiments for this research.
Funding
Open access funding provided by Eötvös Loránd University. This work was financed by grants ÚNKP-19-3 [ÚNKP-19-3-II-ELTE-1009] and GINOP-2.3.2 [GINOP-2.3.2-15-2016-00029].
Author information
Authors and Affiliations
Contributions
TJ and SR supervised the experiments; GB and KB designed the experiments, analysed the data and wrote the manuscript, with contributions from all the authors; GB and KB contributed equally to this work; GB agrees to serve as the author responsible for contact and ensuring communication.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest exists in the submission of this manuscript. The authors declare that the work described has not been published previously, and is not under consideration for publication elsewhere, in whole or in part.
Consent for publication
The MANUSCRIPT has been approved for publication by all authors. All the authors listed have approved the manuscript enclosed herewith.
Additional information
Handling Author: Peter Poor.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Balassa, G., Balassa, K., Janda, T. et al. Expression Pattern of RNA Interference Genes During Drought Stress and MDMV Infection in Maize. J Plant Growth Regul 41, 2048–2058 (2022). https://doi.org/10.1007/s00344-022-10651-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-022-10651-z