Question about the Hi-C heatmap
372 views
Skip to first unread message
PengY
Dec 7, 2021, 5:09:42 AM12/7/21
to 3D Genomics
Hello,
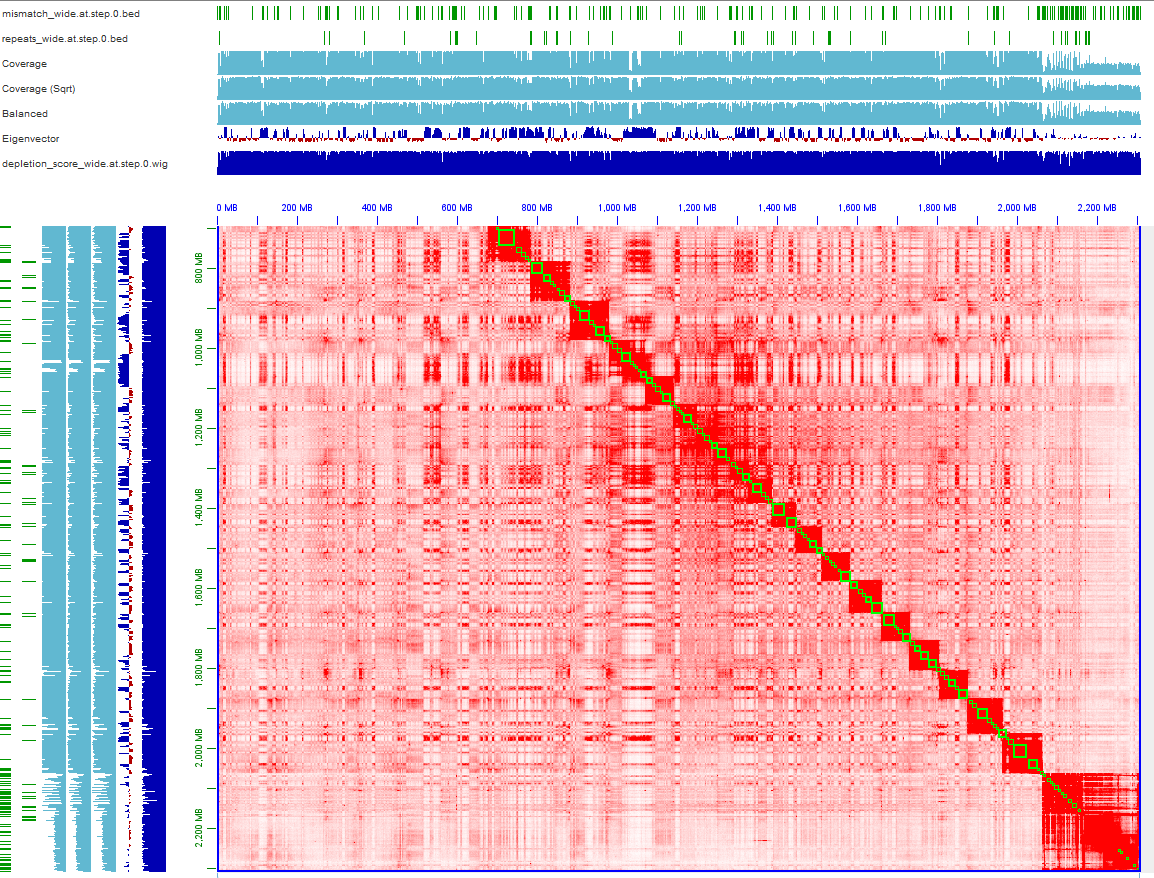
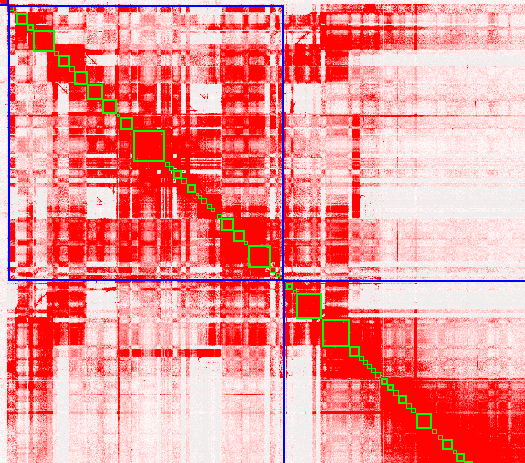
Thanks for your contribution to the great tool 3D-DNA. Recently I asembled one male mammal assembly using Hifiasm and purge_haplotigs. The 3D-DNA pipeline was used for scaffolding draft assembly (>50x Hi-C data). I found there has one strange part in my 0.hic as showing in the below picture. Whole heatmap looks good except the last part which shows obvious coverage and balanced bias. The mismatches are also rich in this region, may due to this region is belong to sex chromosomes. Is it normal? Should I change the parameters such as --editor-saturation-centile 10 or --editor-repeat-coverage? The second picture shows first part of this region, this chromosome looks mess up, corresponding to the chaotic coverage, may due to the high heterozygosis (~2%). Do you think which causes this case? Is there some ways to resolve it?
Looking forward to your reply.


Olga Dudchenko
Dec 7, 2021, 10:17:45 PM12/7/21
to 3D Genomics
Hi Peng,
Looks like for one of your chromosomes the sequences for the two haplotypes are more divergent than for all the others. (Indeed I would expect that would be the sex chromosome, and it's probably relatively young evolutionary speaking). As a result, your contigging/purging procedure ends up being confused, reporting some sequences ones for both haplotypes and some twice, so that you have a mishmash of alt haplotypes that are clashI ing due to collapsed sequences. I would suggest trying to solve this on the contigging side, although if you want a substantial chunk of Y resolved that would sort of defeat the purpose.
Hope this helps,
Olga
PengY
Dec 15, 2021, 11:54:16 AM12/15/21
to 3D Genomics
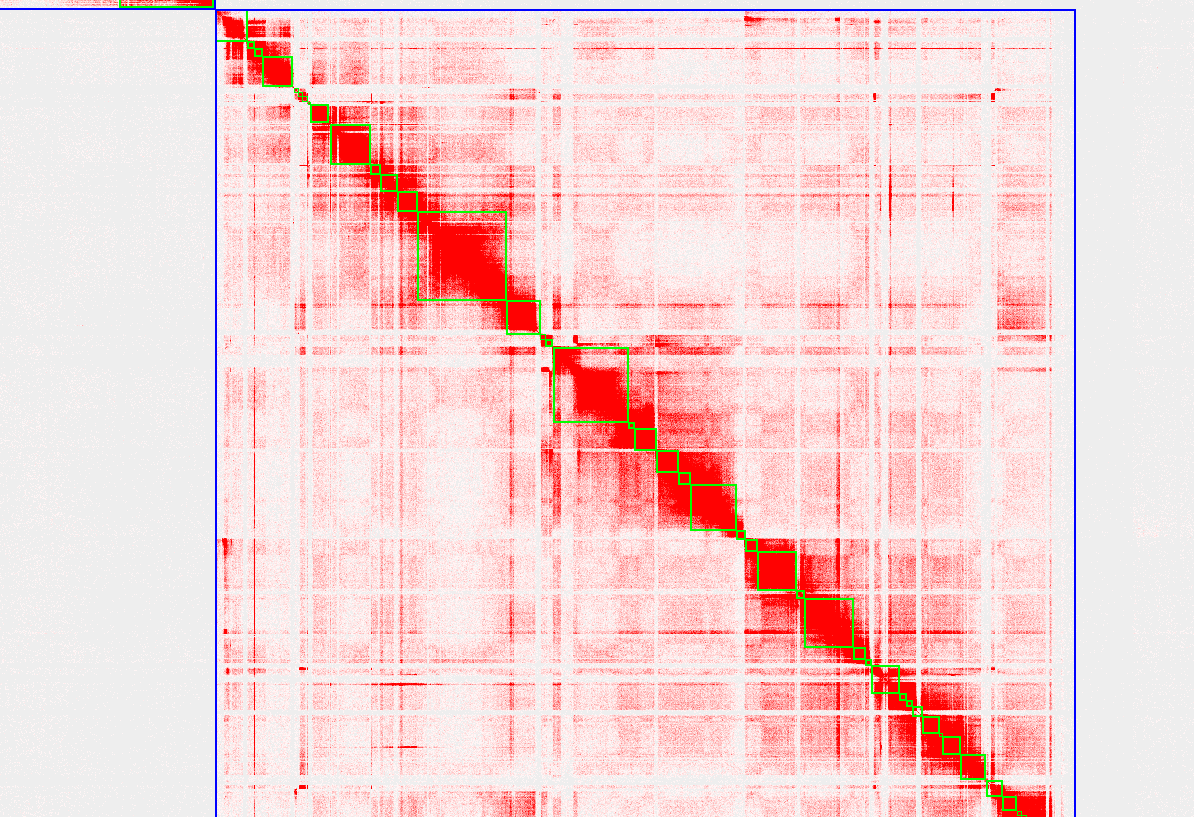
Thanks for your prompt reply. You're right, things become clear when I phased the genome to haplotigs1 and haplotig2 using Hifiasm combined with Hi-C, the first and second picture shows the whole haplotig2 Hi-C heatmap and the Hi-C heatmap of the first chromosome, do you think this chromosome is good enough for being the final version chromosome? Or needs more effort on contigging/purging procedure? The third picture shows the Hi-C heatmap of one chromosome in haplotig1, it looks like lots of alternative haplotypes still remained in the haplotig1, right? How should I deal with them? Could you please give me some suggestions?
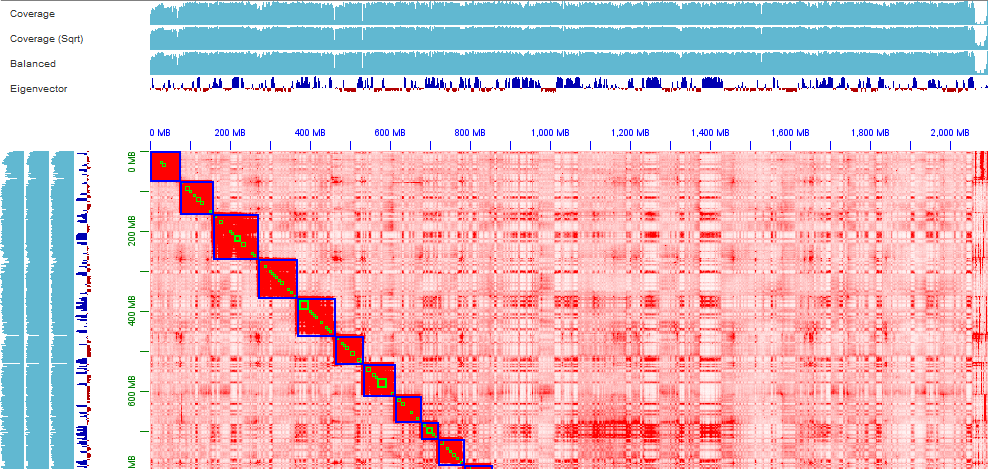
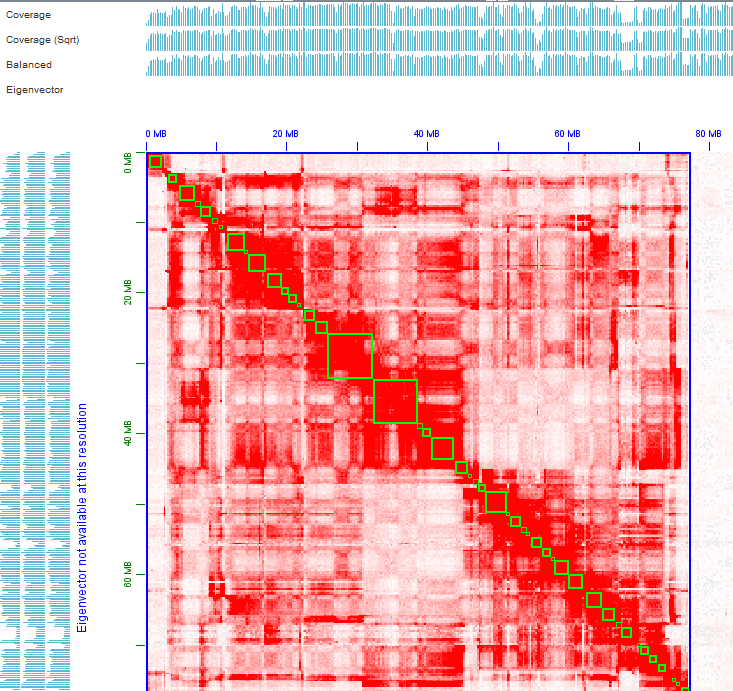



Olga Dudchenko
Jan 7, 2022, 11:05:10 AM1/7/22
to 3D Genomics
I'm sorry I do not understand what I am looking at exactly. But the maps don't look very good to me. -Olga
PengY
Apr 3, 2022, 3:25:41 PM4/3/22
to 3D Genomics
Hello, Olga
I extracted all HiFi raw reads (BTW, we only have totally ~20x coverage HiFi sequencing reads) which come from this part and re-assembled them by more refined pipeline (Briefly, phase the collapsed assembly by SNPs from potential paternal and maternal reads and reassign Hi-C reads on haplotigs1 and haplotigs2, then scaffold them respectively). But the Hi-C maps still don't look good.
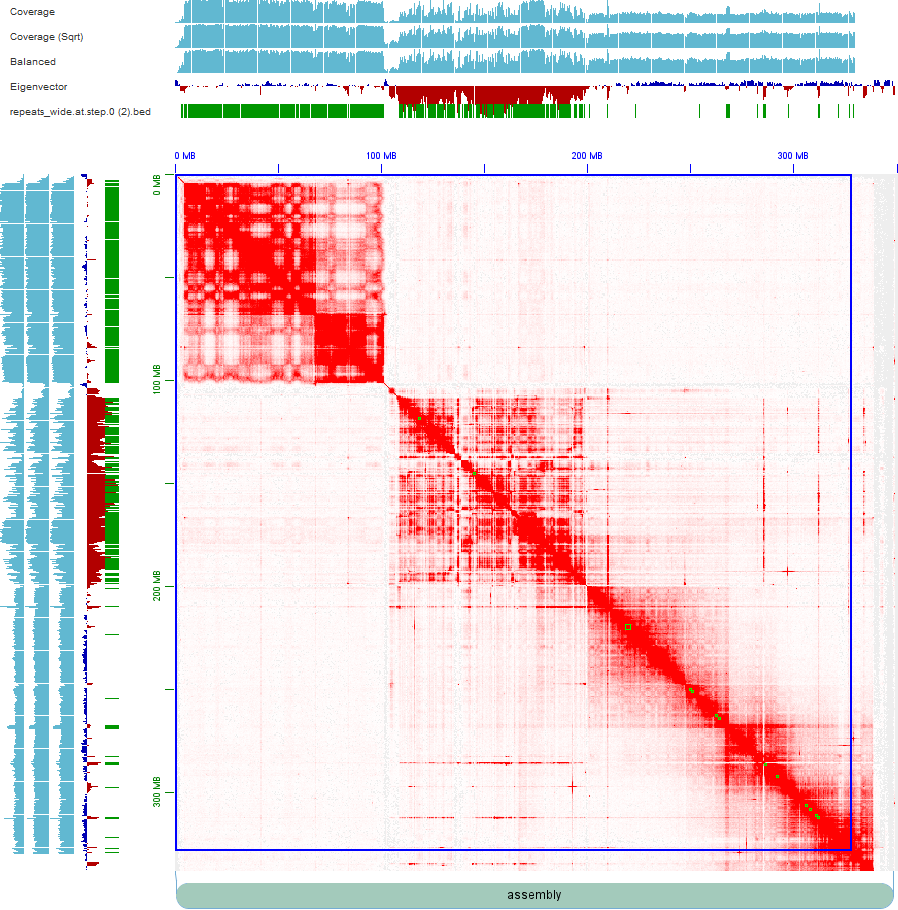
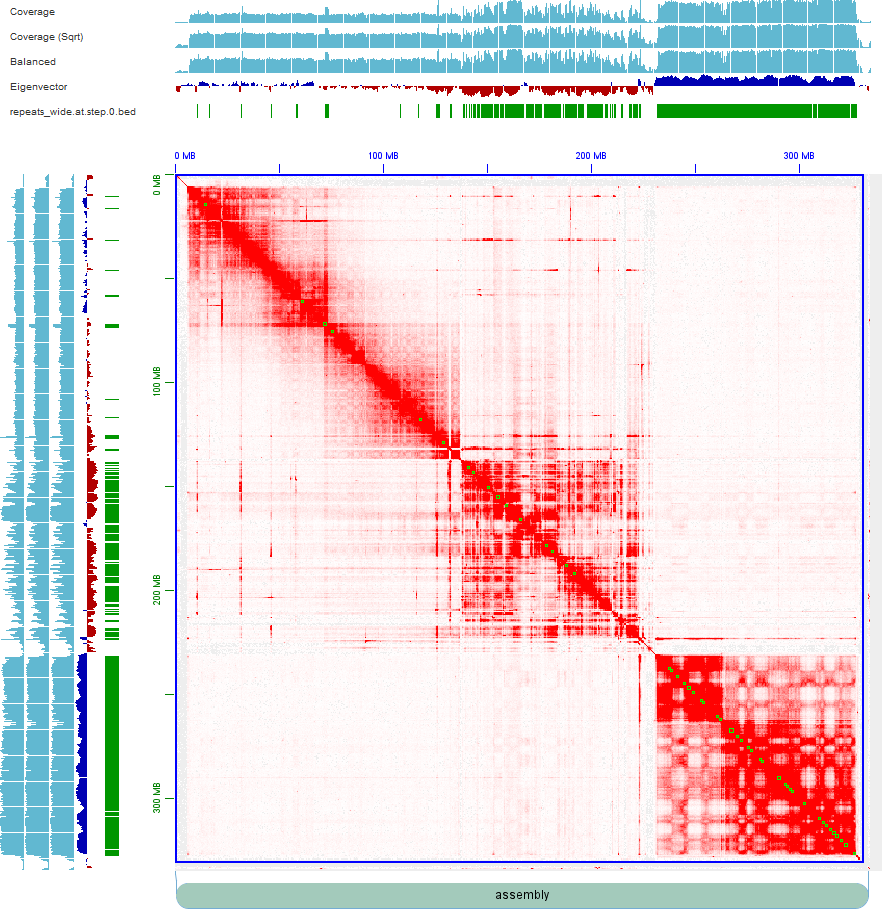
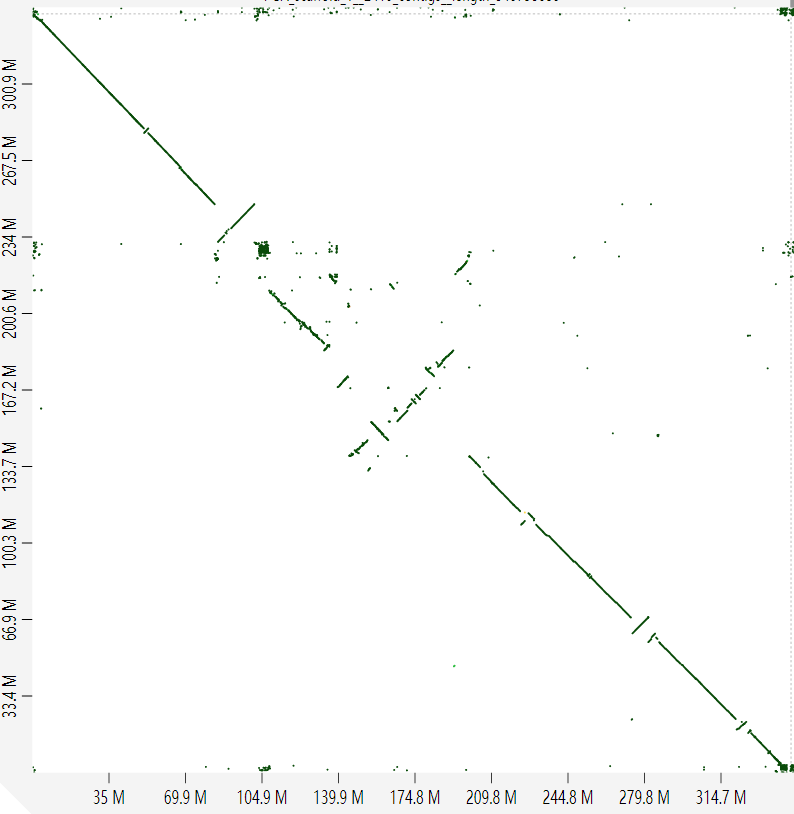
I extracted all HiFi raw reads (BTW, we only have totally ~20x coverage HiFi sequencing reads) which come from this part and re-assembled them by more refined pipeline (Briefly, phase the collapsed assembly by SNPs from potential paternal and maternal reads and reassign Hi-C reads on haplotigs1 and haplotigs2, then scaffold them respectively). But the Hi-C maps still don't look good.


The first picture shows the map of haplotig1, with an autochromosome in the upper left corner and an ancestral X chromosome in the lower right corner. The middle part is more like a young sex chromosome. The second picture (haplotig2) is similar to that of the haplotig1. Collinearity of the two haplotypes indicates that there may still be problems with the middle part of this assembly.

Several contiggers such as Hifiasm, HiCanu, Flye along with different level purge didn't work for phasing this part. Previous study which using male-specific SNPs to phase the young sex chromosome may not work for our species cause both sexes have one pair of this young sex chromosome. Could you please give me some suggestions? I will really appreciate it if you can help me.
Olga Dudchenko
Apr 19, 2022, 1:06:13 AM4/19/22
to 3D Genomics
Have you looked into 3d-dna phasing? See Hoencamp et al. 2021 supplement for details on how the tool works and what it can offer. Perhaps it could help you separate the signal from the young sex chromosomes for the collapsed contigs. Best, -Olga
Reply all
Reply to author
Forward
0 new messages
